Link to online paper: https://academic.oup.com/femsec/article-lookup/doi/10.1111/j.1574-6941.2011.01189.x
Abstract
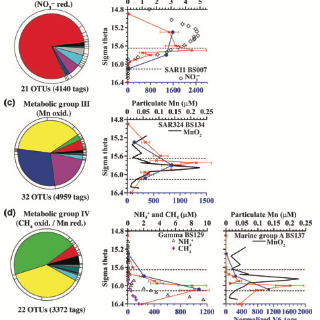
The Black Sea is a permanently anoxic basin with a well-defined redox gradient. We combine environmental 16S rRNA gene data from clone libraries, terminal restriction fragment length polymorphisms, and V6 hypervariable region pyrosequences to provide the most detailed bacterial survey to date. Furthermore, this data set is informed by comprehensive geochemical data; using this combination of information, we put forward testable hypotheses regarding possible metabolisms of uncultured bacteria from the Black Sea’s suboxic zone (microaerophily, nitrate reduction, manganese cycling, and oxidation of methane, ammonium, and sulfide). Dominant bacteria in the upper suboxic zone included members of the SAR11, SAR324, and Microthrix groups and in the deep suboxic zone included members of BS-GSO-2, Marine Group A, and SUP05. A particulate fraction (30 lm filter) was used to distinguish between free-living and aggregate-attached communities in the suboxic zone. The particulate fraction contained greater diversity of V6 tag sequences than the bulk water samples. Lentisphaera, Epsilonproteobacteria, WS3, Planctomycetes, and Deltaproteobacteria were enriched in the particulate fraction, whereas SAR11 relatives dominated the free-living fraction. On the basis of the bacterial assemblages and simple modeling, we find that in suboxic waters, the interior of sinking aggregates potentially support manganese reduction, sulfate reduction, and sulfur oxidation.